Microhomology-Predictor
Microhomology-based choice of Cas9 nuclease target sites.
When programmable nucleases (such as ZFNs, TALENs, or CRISPR) are treated, 1~3bp deletion or 1bp insertion would be induced via nonhomologous end-joining (NHEJ) repair pathway, or deletions using microhomologies of more than 2 bases would be frequently introduced via microhomology-mediated end joining (MMEJ) pathway for repairing double-strand breaks in DNA. With Microhomology-Predictor, one can simply predict the mutation patterns caused by MMEJ pathway and estimate how frequently unwanted in-frame deletions would be happened.
Citation info: Bae S. et al. Microhomology-based choice of Cas9 nuclease target sites. Nature Methods 11, 705-706 (2014).
- Pattern score: score of each pattern according to the microhomology size and the deletion length
- Microhomology score: the sum of all the pattern scores
- Out-of-frame score: the ratio of out-of-frame pattern scores over the sum of all the pattern scores
To avoid unwanted in-frame deletions in a protein-coding sequence as much as possible,
one should choose target sites with high out-of-frame scores. A score above 66 is recommended.
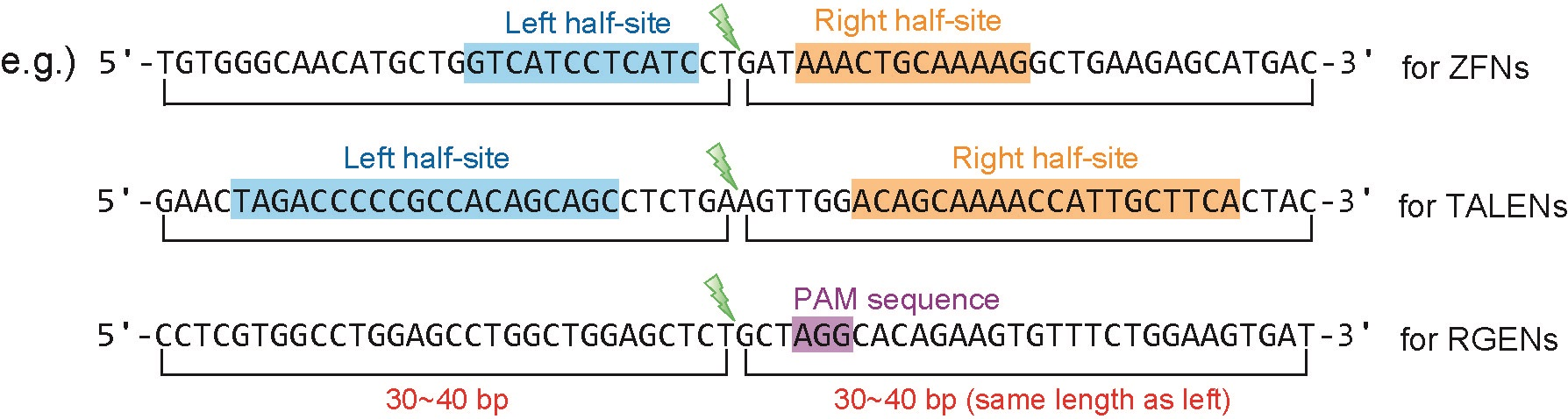
- Calculating microhomology-associated scores for all engineered nucleases [ZFNs, TALENs, RGENs (Cas9 RNA-guided endonucleases), etc.]