About
RGENs, Cas-OFFinder, Microhomology-Predictor, Cas-Designer, and Cas-Database.
RGENs (RNA-guided endonucleases)
In January 2013, several groups (Cho et al, Nat. Biotechnol. 2013; Cong et al, Science 2013; Mali et al, Science 2013; Hwang et al, Nat. Biotechnol. 2013; Jiang et al, Nat. Biotechnol. 2013; Jinek et al, Elife 2013) independently reported a new class of genome editing nucleases — termed RNA-guided endonucleases (RGENs) herein to avoid confusion with the original type II clustered regularly interspaced short palindromic repeat (CRISPR)–Cas (CRISPR-associated) adaptive immune system in bacteria — the specificity of which is mostly determined by small guide RNAs rather than by DNA-binding proteins. They cleave chromosomal DNA in a site-specific manner, which triggers endogenous DNA repair systems that result in targeted genome modification.
An RGEN is comprised of CRISPR -associated protein 9(Cas9), a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA), which formthe dualRNA–Cas9. Alternatively, an RGEN can contain Cas9 and a single-chain guide RNA (sgRNA). The guide sequence in the crRNA (fig a) or sgRNA (fig b) is complementary to a 20bp target DNA sequence known as protospacer, which is next to the 5'-NGG-3' (where N represents any nucleotide) sequence known as protospacer adjacent motif (PAM) (Kim et al, Nat. Rev. Genet. 2014).
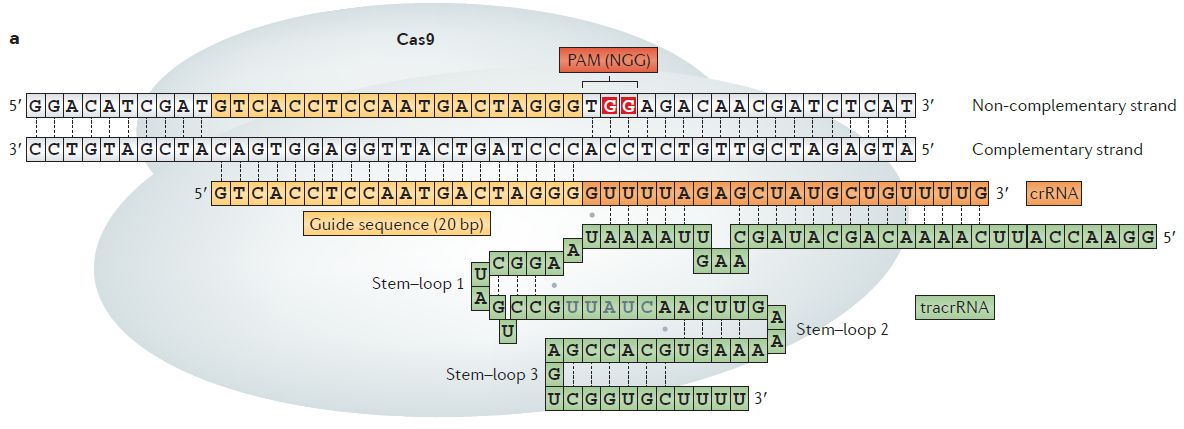
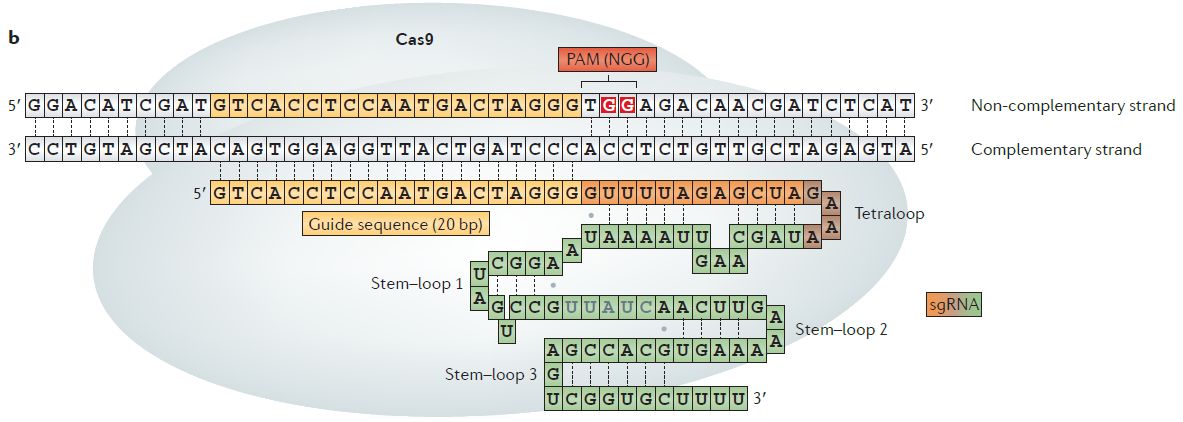
Cas-OFFinder (potential off-target finder for RGENs)
Unfortunately, RGENs cleave not only on-target sites but also off-target sites that differ by up to several nucleotides from the on-target sites (Cho et al, Genome Res. 2014; Fu et al, Nat. Biotechnol. 2013; Hsu et al, Nat. Biotechnol. 2013), causing unwanted off-target mutations and chromosomal rearrangements. To address this issue, researchers must be able to search for potential off-target sites in the genome. Here we introduce a fast and highly versatile off-target searching tool, Cas-OFFinder. Unlike other algorithms currently available for identification of RGEN off-target sites, Cas-OFFinder is not limited by the number of mismatches and allows variations in PAM sequences recognized by Cas9. Cas-OFFinder is available as a standalone command-line program or directly usable via our website.
** Update history
(2014/12/15) - DNA and RNA bulges are allowed for crRNA
(2015/02/23) - Web site is updated
(2015/04) - SaCas9 is added to PAM list (Ran et al, Nature 2015, doi:10.1038/nature14299)
(2015/09/30) - Cpf1 is added to PAM list (Zetsche et al, Cell 2015, doi:10.1016/j.cell.2015.09.038)
(2016/07/06) - Corrected a bug regarding miscalculated position of Cpf1 offtarget sites
(2016/08/18) - Updated to Cas-OFFinder 2.4
Microhomology-Predictor (microhomology-associated score calculator)
Programmable nucleases, ZFN, TALEN and RGENs enable gene knockout in cultured cells and organisms by producing site-specific DNA double-strand breaks, whose repair via error-prone non-homologous end joining (NHEJ) or microhomology-mediated end joining (MMEJ) gives rise to frameshift mutations. In MMEJ pathway, we achieve efficient gene disruption in human cell lines and animals by developing a computer program that assists the choice of nuclease target sites based on microhomology prediction.
** Update history
(2015/02/23) - Web site is updated
Citation info: Bae, S. et al. Microhomology-based choice of Cas9 nuclease target sites. Nature Methods 11, 705-706 (2014)
Cas-Designer (guide RNA design tool for RGENs)
If one researcher would like to search for RGEN targets with low potential off-target effects and high knock-out efficiencies in exon, Cas-designer is very useful. It provides all RGEN targets with the information of potential off-target numbers and out-of-frame scores via Cas-OFFinder and Microhomology predictor.
Please note that mismatch number is fixed to be within 2nt, and optional DNA/RNA bulge is allowed to be within 3nt.
** Update history
(2015/04) - SaCas9 is added to PAM list (Ran et al, Nature 2015, doi:10.1038/nature14299)
(2015/09/30) - Cpf1 is added to PAM list (Zetsche et al, Cell 2015, doi:10.1016/j.cell.2015.09.038)
Cas-Database (genome-wide guide RNA library for spCas9)
Cas9 nucleases from Streptococcus pyogenes (SpCas9) have been widely used for both gene knockout and knock-in at the level of single or multiple genes. Recently, a few groups have undertaken genome-wide Cas9-mediated genetic screens in human and other mammalian cells, but single guide RNA (sgRNA) selection at this scale is difficult.
Cas-Database contains all available SpCas9 target sites in coding sequence regions with several characteristics of each target throughout the genome of 12 different organisms. With an easy-to-use web interface, Cas-Database allows users to select optimal target sequences simply by changing the filtering conditions. Furthermore, it provides a novel and powerful way to select multiple optimal target sequences from hundreds or thousands of genes at once for the creation of a genome-wide library.
Cas-Analyzer (JavaScript-based instant assessment tool for genome edited cell sequencing data)
Cas-Analyzer is an instant assessment tool for high-throughput sequencing data for genome edited cells. It is constructed with a JavaScript-based algorithm; thus, it wholly runs on the client-side so that large amounts of sequencing data do not need to be uploaded to the server. Currently, Cas-Analyzer supports a variety of nucleases, including single nucleases (SpCas9, StCas9, NmCas9, SaCas9, CjCas9, and AsCpf1/LbCpf1) and paired nucleases (ZFNs, TALENs, Cas9 nickases, and dCas9-FokI nucleases).
** Update history
(2016/11) Cas-Database is updated based on Ensembl 86.
(2016/12/14) Opened Cas-Database for Cpf1.