Digenome-sequencing
Genome-wide profiling of off-target effects.
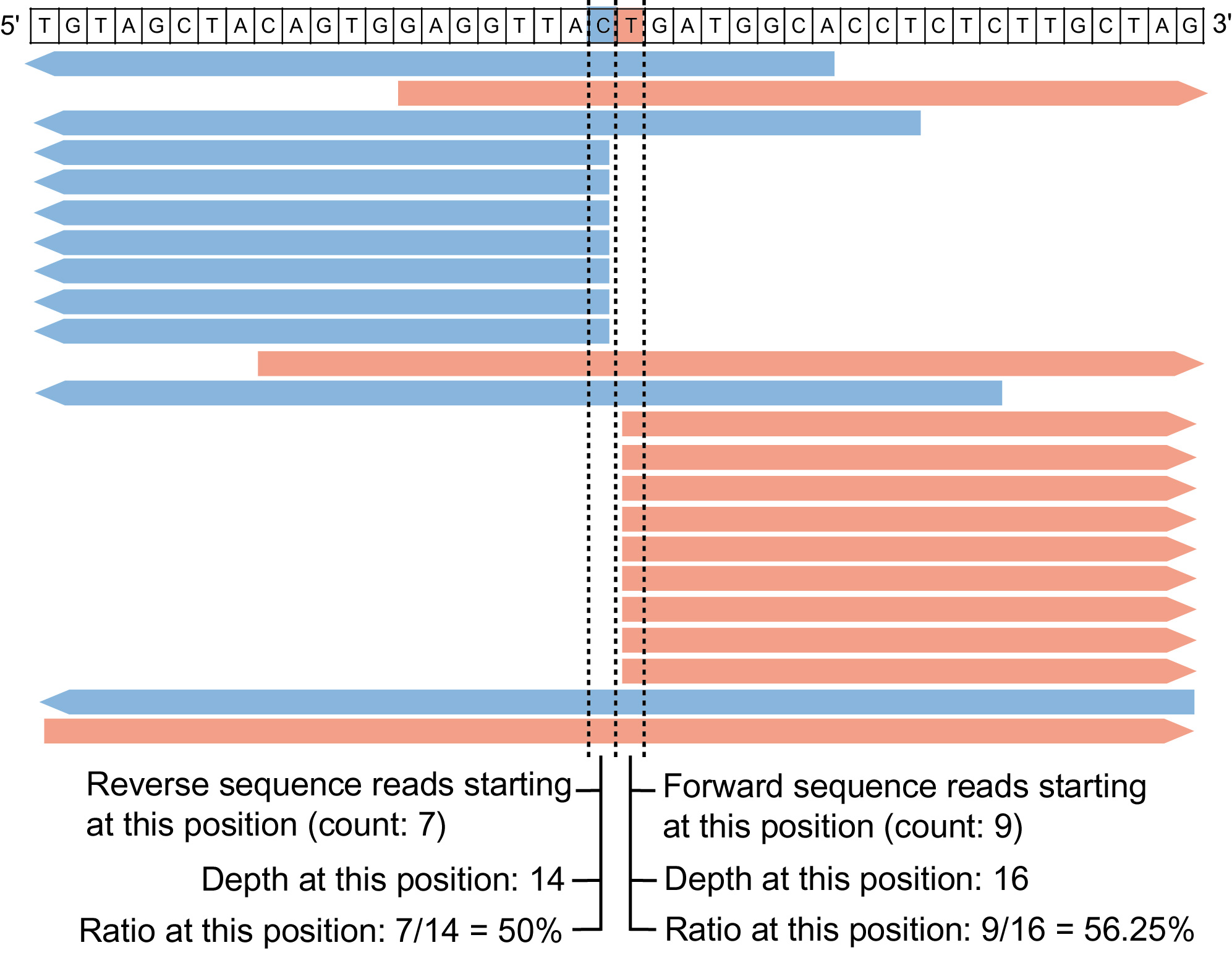
Digenome-seq is an in vitro nuclease-digested whole-genome sequencing to profile genome-wide nuclease off-target effects in cells. This in vitro digest yields sequence reads with the same 5' ends at cleavage sites that can be computationally identified by Digenome-seq program. Thanks to Emscripten, this web version of Digenome-seq program completely runs on the client-side so that large amounts of sequencing data do not need to be uploaded to the server.
Citation info:
Please input your data in below form, or you can download a standalone version of this tool here. Also you can try this tool with an example data. Please check example protocol of Digenome-seq experiment in help page.
Cleavage type
e.g.) Cas9: blunt end, AsCpf1, LbCpf1: 3 overhang (5'), FnCpf1: 5 overhang (5'), ZFN: 4 overhang (5'), TALEN: 6 overhang (5')
Target sequence(s), one sequence per line (optional, 5' to 3'):
Minimum mapping quality for bam reads
Minimum number of forward reads with same 5' ends
Minimum number of reverse reads with same 3' ends
Minimum depth at each position
Minimum ratio at each position
Minimum cleavage score [?]

